TUM bioinformaticists shed light on the structures of dark proteins
The "dark matter" in the protein universe
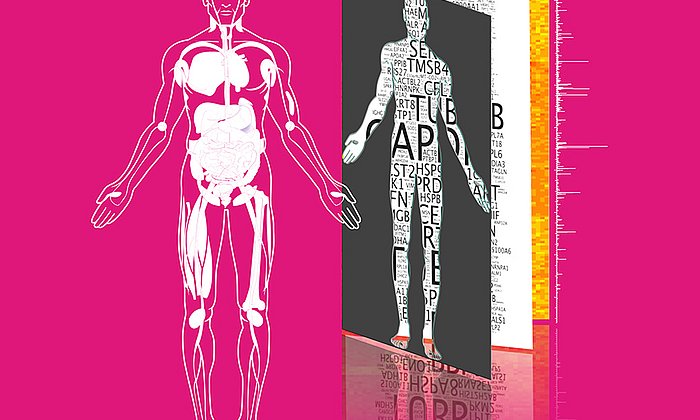
Fifteen percent of the mass of an average human: that is the overall amount of all proteins, also known as the proteome. The protein molecules perform essential functions in the body and cells. They initiate metabolic processes, help the organism ward off diseases and ensure that vital biological substances are transported.
The function of these proteins is significantly determined by their three-dimensional structure. Yet there are also proteins that either completely or at least partially differ from the structure that has so far been elucidated by scientific experiments. As a result, their structure cannot be modeled. Researchers refer to these proteins and protein building blocks as "dark proteins" and collectively as the "dark proteome", in analogy to the dark matter found in outer space. Up until now, scientists still did not know how many of these proteins form part of the dark proteome.
Half of the proteome is dark
Together with the Commonwealth Scientific and Industrial Research Organisation (CSIRO) in Sydney and the University of Lisbon, Dr. Andrea Schafferhans from the Chair of Bioinformatics at TUM studied the properties of the dark proteome. They did so by filtering out pieces of information from various databases, linking them with each other and evaluating the data.
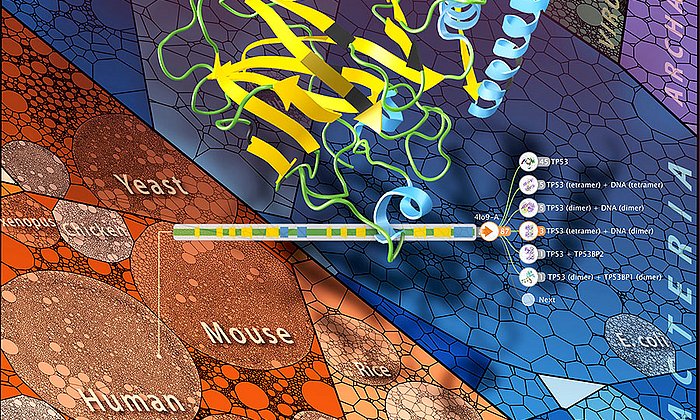
The "Aquaria" database, a joint project organized by CSIRO and TUM, played an important role in this process. The website went live in early 2015 and allows all researchers to have the 3D structure of protein sequences computed. The database does this by referring to existing structures and using that information to create a probable model for the new structure. This website helps researchers determine which protein structures are in fact "dark".
The result: Half of the proteome of all living beings whose cells have a nucleus – and that includes humans – is part of the dark proteome. "And again half of that has structures that remain completely unidentified," says Schafferhans.
Few relatives, hardly any interactions with other proteins
Furthermore, the researchers were able to identify the following properties of dark proteins: Most dark proteins are short, rarely interact with other proteins, are frequently excreted and only have a small number of evolutionary relatives.
The scientists also found out that some of the assumptions previously held about dark proteins are in fact wrong. The majority of them are not disordered proteins, for example. The latter only adopt their actual structure once they perform a function. The remainer of the time they aopt varying shapes. Moreover, the majority of dark proteins are not transmembrane proteins, i.e. proteins located inside a membrane that separates cell components or entire cells from each other. Up until now, these two assumptions were believed to explain why the structure of dark proteins is so hard to identify.
With their findings, which were published in the specialist journal Proceedings of the National Academy of Sciences, the researchers have laid important groundwork that will help scientists better analyze these mysterious protein molecules in the future.
The researchers also hope to draw more attention to the dark proteome, because it could contain proteins that play a key role in human health.
Medical protein research at TUM
Biomedicine is a key research area at TUM, which consequently combines basic and applied research. As part of this concept, the following new research facilities were set up: the TUM Center for Functional Protein Assemblies (CPA), the Bavarian Nuclear Magnetic Resonance Center, the Central Institute for Translational Cancer Research of the Technical University of Munich (TranslaTUM) and the Klaus Tschira Foundation’s Research Center for Multiple Sclerosis. As an integrative research center, TUM’s MUNICH SCHOOL OF BIOENGINEERING forms a joint learning and research platform for all pertinent activities carried out in medically relevant fields of engineering in the various faculties, including imaging technologies. TUM is also a major stakeholder in the Center for Integrated Protein Science Munich (CIPSM) cluster of excellence.
Publication:
Nelson Perdigãoa et al.: Unexpected features of the dark proteome. Proceedings of the National Academy of Sciences (2015). DOI: 10.1073/pnas.1508380112
Link: www.pnas.org/content/early/2015/11/16/1508380112
Contact:
Dr. Andrea Schafferhans
Technical University of Munich
Chair of Bioinformatics, Prof. Burkhard Rost
Tel.: +49 289 17833
andrea.schafferhans@rostlab.org
Technical University of Munich
Corporate Communications Center
- Stefanie Reiffert
- reiffert@zv.tum.de
- presse@tum.de
- Teamwebsite