Study establishes the first public collection of bacteria from the intestine of mice
Mouse gut bacteria find a new home
They are microscopically small and live both on humans and animals. They can help with recovery from an illness or literally make you sick: Billions of micro-organisms, most of which are found in the intestines, as well as on the skin and other regions of the body, living in symbiosis with the host. These tiny beings are of central importance, and experts refer to them as intestinal microbiota or the microbiome. Decoding its characteristics and obtaining a better understanding of it is what scientists at the Central Institute for Nutrition and Food Research (ZIEL) at the Technical University of Munich (TUM) are working on.
76 cultured bacterial species from the mouse microbiome identified and archived
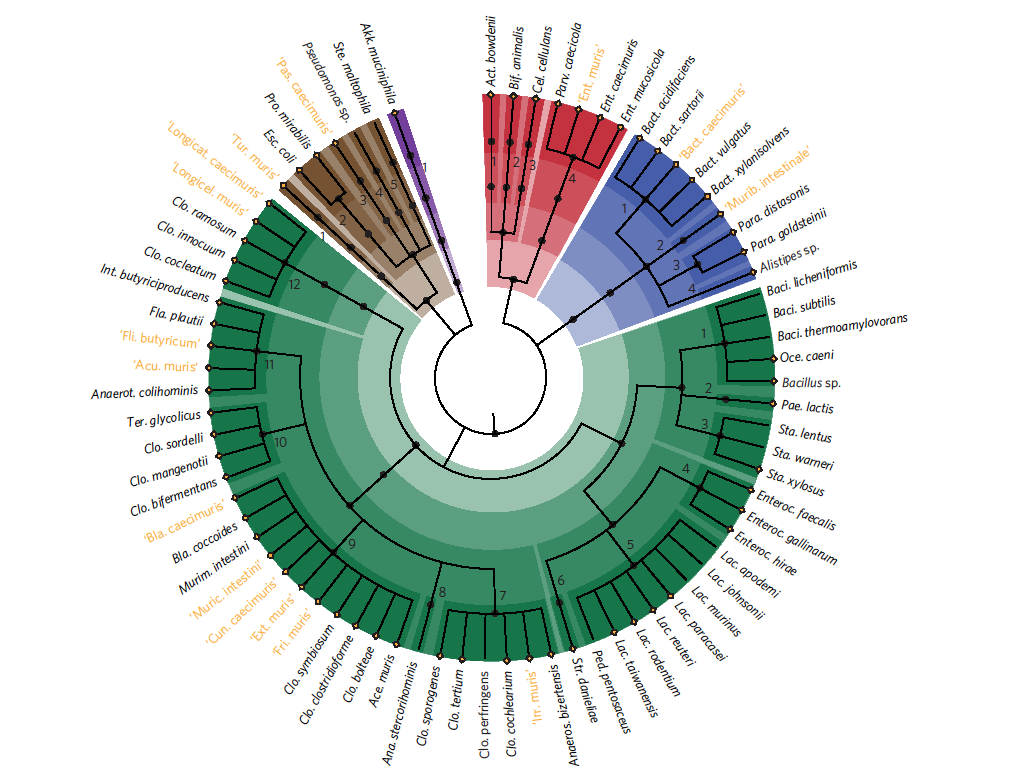
One key to obtaining information about the interactions between gut bacteria and their host are mouse models. However only a handful of mouse intestinal bacteria have been made publicly available and fully characterized so far. This is a highly limiting factor for research, because it complicates the annotation of data obtained by molecular techniques, and because it has been shown that gut microbiomes are to some extent specific to their host, and researchers have been using strains of other origin in mouse models. Dr. habil. Thomas Clavel from ZIEL and colleagues describe a new resource in "Nature Microbiology" which, for the first time, contains a hundred cultured bacterial strains from the mouse gut microbiome. For this study, 1500 cultures were examined, and 76 different species were identified and archived.
"The goal of our work was to take a big initial step towards decoding the cultured fraction of gut bacterial communities in mice. There is still a lot left to do. We will be making our work available to scientists around the world and hope that others will also help to find the pieces to complete the puzzle", said Clavel, who has been researching various bacteria in gut microbiomes at the TU Munich for ten years. Although the mouse gut microbiome presents a number of similarities with the human microbiome, the work showed that around 20 percent of the strains in the collection prefer colonizing the intestines of mice.
In order to better understand colonization processes in the intestine, bacteria first need to be identified and characterized in detail. "Because mouse models are indispensable for preclinical studies, the resource now made available shall contribute to a better understanding of microbe-host interactions and to a higher degree of standardization", said Clavel.
New bacteria with specific functions
For the first time, the researchers were able to characterize new bacteria with important functional properties: For example Flintibacter butyricum produces the short-chain fatty acid butyrate from both sugars and proteins—a rare property in the realm of intestinal bacteria. Butyrate is a main product of fermentation in the intestine, and has been shown to have anti-inflammatory and positive effects against metabolic diseases in numerous studies.
"We still have a lot of gaps in our knowledge about gut microbiomes, but with the publicly available database of cultured mouse gut bacteria and their genetic material, we are now a little closer to our goal", Thomas Clavel from the TUM stated enthusiastically.
Publication:
Ilias Lagkouvardos, Rüdiger Pukall, Birte Abt, Bärbel U. Foesel, Jan P. Meier-Kolthoff, Neeraj Kumar, Anne Bresciani, Inés Martínez, Sarah Just, Caroline Ziegler, Sandrine Brugiroux, Debora Garzetti, Mareike Wenning, Thi P. N. Bui, JunWang, Floor Hugenholtz, Caroline M. Plugge, Daniel A. Peterson, MathiasW. Hornef, John F. Baines, Hauke Smidt, Jens Walter, Karsten Kristiansen, Henrik B. Nielsen, Dirk Haller, Jörg Overmann, Bärbel Stecher und Thomas Clavel: A Mouse Intestinal Bacterial Collection (miBC) provides Host-Specific Insight into Cultured Diversity and Functional Potential of the Gut Microbiota, Nature Microbiology 08/2016.
DOI 10.1038/nmicrobiol.2016.131
Contact:
Dr. habil. Thomas Clavel
Technical University of Munich
ZIEL – Institute for Food and Health
Core Facility NGS/Microbiome
Phone: +49 81 61 71 55 34
Mail: thomas.clavel@tum.de