Scientists measure molecular forces between nucleosomes
In the molecular bench vise
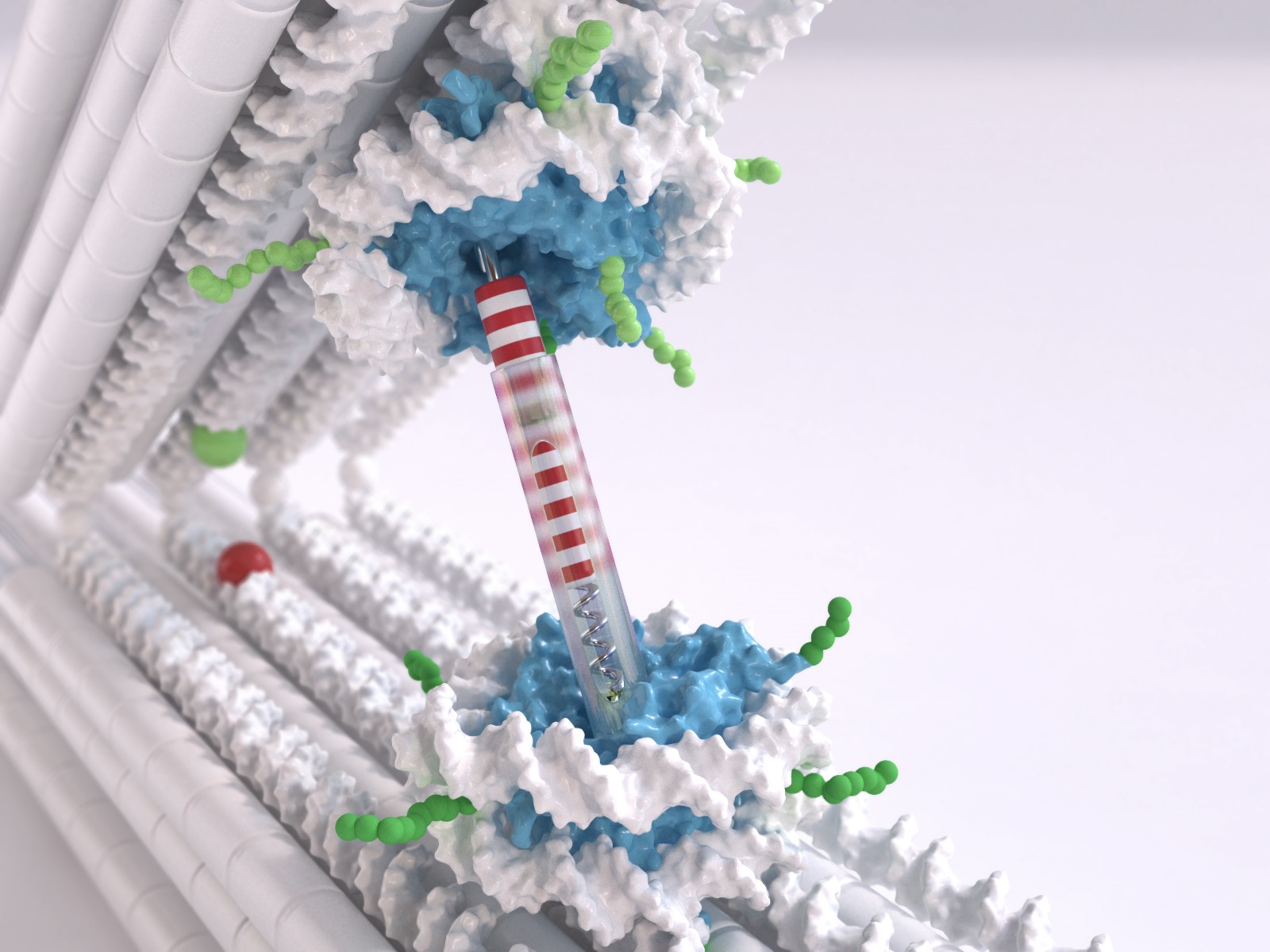
The nucleus of every human cell has to contain two meters of the molecule deoxyribonucleic acid (DNA), the carrier of our genetic information. This means the DNA has to be packaged carefully, primarily wrapping itself around certain proteins. Comparable to small spools, these structures made of DNA and proteins are referred to as nucleosomes. Nucleosomes are linked to one another by segments of DNA that have not been wound up. Viewed under the electron microscope, the DNA packed in nucleosomes resembles beads on a string.
The mutual interaction of the nucleosomes with one another and the resulting higher-order structures have yet to be completely explained. A team of scientists led by Hendrik Dietz of the Technical University of Munich (TUM) and Philipp Korber of the Ludwig-Maximilians-Universität Munich (LMU) has now made a substantial contribution to solving this puzzle: For the first time ever they have succeeded in directly measuring the attractive forces at work between the nucleosomes. Their results have been published in the journals "Science Advanced" and "Nano Letters".
DNA Origami: Integrating nucleosomes in the tweezers
Dietz, holder of the Chair for Experimental Biophysics at the TUM, uses DNA as a construction material to build molecular structures, a technology referred to as DNA Origami. He and his team have now developed a tweezer structure, consisting of two rigid DNA bars connected by a joint, which can be used to measure the interactions between the nucleosomes. One nucleosome structure was integrated in each bar. "We can control the position and orientation of the nucleosomes in the DNA tweezers with a very high degree of precision," says Dietz. "This is very important when it comes to really being able to measure the interactions."
The LMU researchers tackled the challenge of developing nucleosome structures that can be integrated into the tweezers. "Normally the nucleosome has two double-stranded ends of the rolled up DNA very close to one another," Korber explains. "But what we needed were two protruding single strands, more in the middle. This was a significant issue, since it can destabilize the entire structure. Our team member Corinna Lieleg nevertheless succeeded in finding the right spots for these handles."
This meant the researchers were able to measure a very weak interaction between the nucleosomes, with 1.6 kcal/mol at a range of about 6 nanometers.The orientations of the nucleosomes to one another showed hardly any effect. However, certain chemical changes in the nucleosomes further weakened the interactions.
Does the 30 nanometer fiber really exist?
The result could help resolve a current scientific dispute. According to the current theory, nucleosomes together with other proteins form a type of super-spiral with a diameter of 30 nanometers, referred to as the 30 nanometer fiber. Up to now however, this higher-order structure has never been observed in living cells. Whether or not the DNA packaging, the chromatin, really takes on the form of such a super-spiral is at the moment highly controversial. The minute forces between the nucleosomes, which the researchers have now successfully measured, appear to contradict the accepted theory. "Our data point to very soft structures that are easily deformed by external influences," says Dietz. "Although our work can't conclusively resolve the current debate, it's still an important contribution that makes it possible to rule out a couple of potential models."
The keen interest in the higher-order nucleosome structure is understandable, since its form is of fundamental importance. Only those genes which are in a relatively non-compact chromatin structure are "active", meaning that the proteins coded there are actually produced by the cellular machinery.
Gene regulation goes awry within cancer cells
"Over the last ten years it has become clear that many changes and mutations that turn cells into cancer cells take place at this level," Korber says. In a cancer cell the cellular decisions about which genes are active and which are inactive go awry. Genomic regions which should not be accessible are left open and vice versa. "But if it's only the packaging and not the gene itself which is defective, there is a therapeutic hope of being able to revert the packaging again." A cure would be much more difficult if the gene itself was completely deleted from the genome.
The researchers want to apply the molecular tweezers they developed for measuring the forces between nucleosomes in the investigation of other structures. "In biology the orientation of structures to one another is always important," says Korber. "Now we have a kind of molecular bench vise which we can use to specifically control the spatial orientation of structures to one another."
In another experiment the researchers have already measured the force needed to unwrap the DNA from the nucleosome. The researchers were thus able to demonstrate that the measurement system can be used to measure both the forces acting between molecules and the forces acting within the molecules themselves.
Publications:
Funke et al.: "Uncovering the forces between nucleosomes using DNA origami", Science Advanced,
DOI: 10.1126/sciadv.1600974
advances.sciencemag.org/content/2/11/e1600974.full
Funke et al: "Exploring Nucleosome Unwrapping Using DNA Origam"; Nano Lett., 2016, 16 (12), pp 7891–7898; DOI: 10.1021/acs.nanolett.6b04169
pubs.acs.org/doi/abs/10.1021/acs.nanolett.6b04169
Highresolution pictures:
Contact:
Prof. Hendrik Dietz
Technical University of Munich (TUM)
+49 (0) 89 289-11615
dietz@tum.de
bionano.physik.tu-muenchen.de
PD Dr. Philipp Korber
LMU Munich / Ludwig-Maximilians-Universität München
Molecular Biology
Biomedical Center Munich
+49-89-218075435
pkorber@lmu.de