Interaction between tumor suppressor protein and chaperone revealed:
A chaperone for the “guardian of the genome”
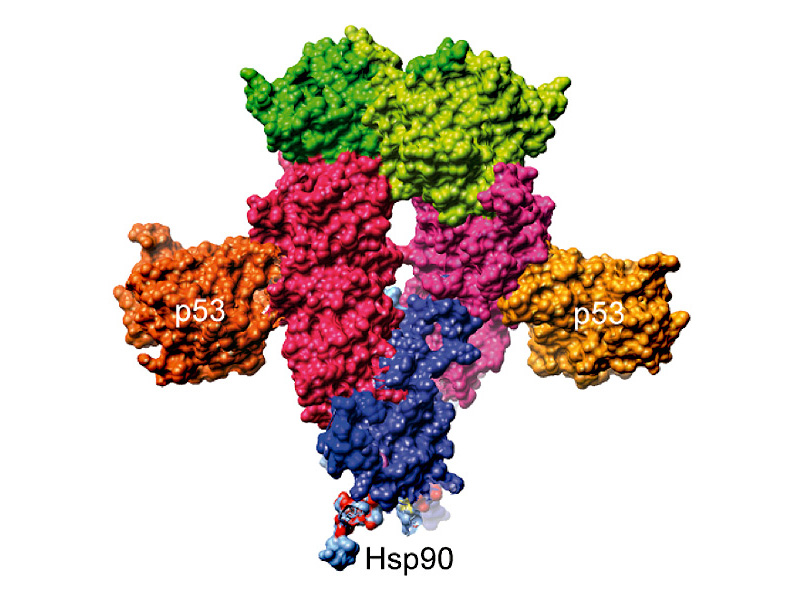
Every cell has thousands of proteins whose activity and lifetime must be regulated to control the cellular life cycle from cell division to cell death. The heat shock protein Hsp90 plays a key role in this process. It is a so-called chaperone, a quality controller, as it were. It monitors and controls the quality and activity of many important signal proteins and helps them take on the right form. When the cell is exposed to high stress levels from heat or a lack of oxygen, Hsp90 is produced in larger quantities to shield its partner proteins from damage.
One of the most important partner proteins of Hsp90 is the tumor suppressor protein p53. It prevents the development of cancer at a number of points in the cell and is thus aptly referred to as the “guardian of the genome”. When a cell’s DNA becomes damaged, p53 ensures that the cell no longer divides and activates repair mechanisms. When too much genetic damage accumulates, the protein initiates a controlled cellular “suicide”. When p53 is inactive, the cell continues to divide in spite of the damage and a tumor develops. In over half of all tumors the p53 protein is damaged or inactivated, meaning the control function cannot be carried out.
Hsp90, on the other had, binds to p53 and keeps it in a functional state until it can fulfill its actual purpose: Binding to specific DNA elements. However, exactly how and where the binding of p53 to Hsp90 takes place was hitherto neither clear nor had it been structurally characterized.
At the Department of Chemistry of the TU München, a team of biochemists headed by Professor Horst Kessler, in collaboration with the group of Professor Johannes Bucher, Chair of the Department of Biotechnology, has now succeeded in working out the details of how p53 binds to Hsp90. Horst Kessler was Chair of the Department of Organic Chemistry and Biochemistry at the TU München from 1989 to 2008 and has been Carl von Linde Professor at the Institute for Advanced Study at the TU München (TUM-IAS) since October 2008.
Using nuclear magnetic resonance (NMR) spectroscopy, the scientists at the Bavarian NMR Center in Garching were able for the first time to characterize the interaction surfaces between Hsp90 and p53 and show that p53 binds to Hsp90 in an already structured form. p53 is thus kept in a functional state until this interaction is terminated by its actual intended binding partner, DNA. To keep p53 in the desired state, a number of interaction surfaces at different sites of the Hsp90 protein must interact in a closely coordinated manner.
In nuclear magnetic resonance spectroscopy, a sample of dissolved proteins is placed in an extremely strong, homogenous magnetic field and exposed to a complex sequence of radio frequency impulses. The atomic nuclei of the protein react to these impulses with a characteristic response frequency that depends on the environment of the respective nucleus. The scientists can measure this response. “Every single stimulated nucleus responds at its own frequency,” explains Kessler. “In this way we can determine the connections between individual nuclei and thus deduce the structure of the protein.” When p53 binds to Hsp90, the response frequencies at specific sites of the protein change. Based on these changes, the scientists can infer the sites at which p53 binds to Hsp90.
The new insight on the interaction surfaces between Hsp90 and p53 are of great significance in the development of new cancer medications. After all, Hsp90 stabilizes not only intact p53, but also, and above all, mutated versions of the protein. This leads to a negative effect of the chaperone. The reason: The defective p53 sustained by Hsp90 binds, in its turn, to an active p53 and inactivates it – potentially leading to a tumor. In the future, medication that acts on the discovered sites could prevent Hsp90 from attaching to and stabilizing defective p53 in cancer cells. “Many of the altered p53 variants in tumors are less stable than intact p53 and require Hsp90 all the more,” explains Franz Hagn, corresponding author of the study. “When this interaction is inhibited, especially mutated p53 is disposed of. This allows the intact p53 to fulfill its function, eliminate damaged cells and prevent the cancer.”
In their study, Kessler, Buchner and their team determined that p53 binds to not only, as previously assumed, to the middle domain of Hsp90, but also with a high affinity to a site in the C terminal domain of the protein. Negatively charged amino acids at both binding sites of Hsp90 are responsible for the stability of this bond. “These sites are similar to DNA, whose phosphate backbone is also negatively charged,” explains Kessler. “So Hsp90 imitates p53’s actual partner. The complex of both proteins is held together in this way.” p53 retains its original state, all the while, allowing it to continue binding to DNA.
This research was supported by the TUM Institute for Advanced Study, the German Research Foundation DFG (SFB 594), the Excellence Cluster Center for Integrated Protein Science Munich (CIPSM), the Bavarian Elite Network, the Fund of the Chemical Industry, and the European Molecular Biology Organization.
Original publication:
Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53 Franz Hagn, Stephan Lagleder, Marco Retzlaff, Julia Rohrberg, Oliver Demmer, Klaus Richter, Johannes Buchner and Horst Kessler Nat. Struct. Mol. Biol., DOI: 10.1038/nsmb.2114
Contact:
Prof. Horst Kessler
Institute for Advanced Study and
Department of Chemistry Technische Universität München
Lichtenbergstr. 4, 85748 Garching, Germany
Tel.: +49 89 289 13300 Fax: +49 89 289 13210
Email: Horst.Kessler@tum.de
http://www.org.chemie.tu-muenchen.de/akkessler/
Prof. Johannes Buchner
Department of Chemistry
Technische Universität München
Lichtenbergstr. 4 D-85748 Garching
Tel.: +49 89 289 13341 Fax: +49 89 289 13345
Email: Johannes.Buchner@tum.de
http://www.biotech.ch.tum.de/